Data can Heal
The developed use-case will be dedicated to the qualitative assessment of cardiac anatomy, based on cardiac imaging, namely the characterization of myocardial wall thinning from cardiac Computed Tomography (CT) images. This descriptor has demonstrated to be a descriptor of interest to diagnose different cardiac diseases and plan the treatment, including: (i) ventricular tachycardia (VT) ablation, to provide surrogates of VT sources that the electrophysiologist has to treat using radiofrequencies ablation; (ii) revascularization in case of coronary artery diseases: to predict function regaining after revascularization. ML methods that will be used to automatically extract myocardium wall boundaries, and thus to compute its
thickness, will be based on Deep learning (DL) techniques, which have demonstrated their ability to segment cardiac images. The main limitation of the widely used DL methods is their lack of generalization capabilities when presented with unseen data, typically images acquired with another imaging system, or even with the same system but configured differently, or corresponding to other pathologies. Another limitation is the need for annotated (e.g. manually delineated) images to train and evaluate the models.
The PAROMA architecture will be the opportunity to go forward towards overcoming these limitations by exploiting the data from different clinical centres. The two partners involved in this use case (UR1 and 6GHI) will be considered as edge platform nodes, each one with its own Local Processing Node. Firstly, two local models will be optimized independently from local images of both centres. It will be based on a 3D U-Net model, which is the most popular model architecture for medical images segmentation. This model will be implemented and trained to segment the inner and outer walls of the left ventricle. Both generated models will be evaluated locally on annotated data using a cross-validation process. This approach will be considered as the baseline.
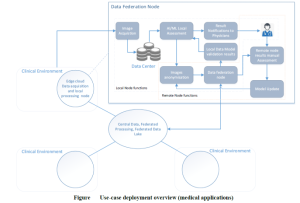
Different scenarios will be implemented to generate models exploiting data from both partners, based on the transmission of the generated partial models to the other edge network node and/or to the central node.
1. Transfer learning with fine-tuning: based on the partial model transmitted by another node, transfer learning will be used to adapt the model to the domain of the local data. The resulting model will be transferred back
to the first node to evaluate the potential improvement/deterioration of the performances on the first dataset.
2. Partial models merging: the partial models from the different nodes will be merged using an ensemble learning strategy. The resulting merged model(s) will be evaluated on both centres.
3. Full federated learning directly exploiting the data from the two centres: the global model will be learned by the central node, by transmitting intermediate models to the edge nodes. In an iterative process, edge nodes will locally optimize the models based on their local data before transmitting them back to the central node.
These scenarios will be jointly implemented by both edge nodes and by the central platform node. They will be evaluated and compared on reference data from both centres to demonstrate the added value of the proposed architecture.
Your Question
Your question is not listed yet? Submit your question with the form below – we will get back to you in due time.

